The quarterly scientific journal
Optimization and Validation of digital PCR assays
Clean Cells, a GMP-certified company, is a world-renown quality control expert with an extensive track-record of qPCR testing of thousands of biologics intended for human or animals.
Following the qualification of our digital PCR platform, the team at Clean Cells worked on the initial optimization and validation of quantitative assays to support existing internal methods. Below is a summary of the work performed within our quality frame to pave the way to a GMP setup which is already made available to customers to support product characterization and release.
This optimization and validation work also shows our team’s ability to develop and validate customized assay from the ground up for sponsors with specific characterization requirements. Validation of method and equipment is performed within our QMS and follows relevant international guidelines, including ICHQ2(R1) and 21CFR part 11.
Method and capabilities
The method
Digital PCR represents an undeniable quality leap, using physical partitioning and automation to grant high quality and robust results. In this straightforward approach, samples are first diluted, then partitioned, amplified before undergoing read-out , resulting in reduced operator time and thus increasing reliability.
Digital PCR may be used in diverse settings to evaluate products requiring increased sensitivity and absolute quantification with a low limit. It can be used independently for research purposes or complementary to TCID50 and FISH methods and to clonality studies (number of transgene copies) in a regulatory setting.
Capabilities and advantages
At Clean Cells, digital PCR can be used for various purposes:
- Gene or plasmid copy number (GCN, PCN)
- Titration (viral vectors, virus seed stocks)
- Quantification of translocations in modified cells
- Gene expression
- Host cell DNA quantification
Clean Cells implemented a digital PCR platform which presented several advantages when developing, optimizing, validating and executing dPCR methods:
- 5 simultaneous quantifications in the same well thanks to the five fluorescence channels available on our platform QIAcuity One 5plex from QIAGEN.
- Same efficiency of development and validation as for qPCR
Optimization and validation
Optimization work
Optimization of dPCR assays can be performed on different parameters such as primers and probes concentrations, the annealing temperature or the number of amplification cycles, with the same objectives: improving the discrimination between positive and negative partitions and reducing the rain phenomenon.
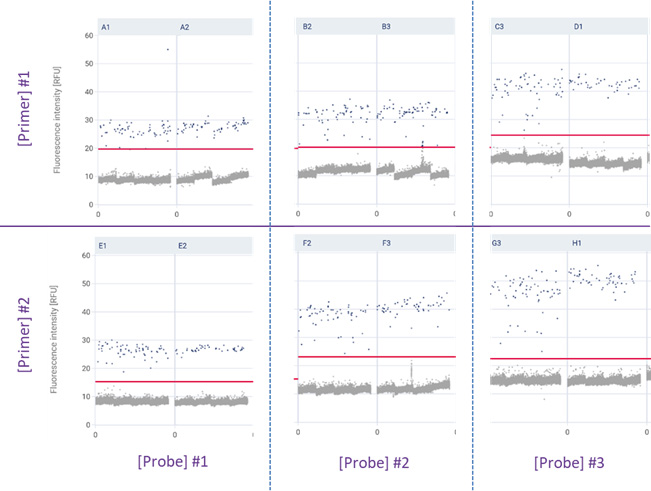
Example of results for primers and probe concentrations for a duplex assay:
The data shows that there was no significant difference between the two tested primer concentrations, but the increase of probe concentration improves the discrimination between positive and negative partitions.
Validation of assays
A duplex assay aims to measure the expression of three target genes in eukaryotic cells by quantifying in the same well a housekeeping gene and the target. After optimization, the three assays have been validated according to ICHQ2(R1) guideline with the evaluation of precision, specificity, linearity, range, accuracy and robustness.
Accuracy
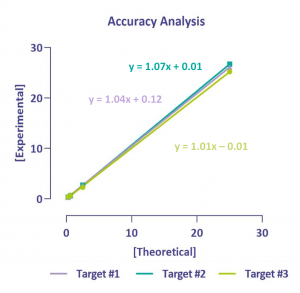
The quantification of these three targets was very accurate with a high sensitivity allowing quantification of target expressed as low as 0.2%
Sensitivity
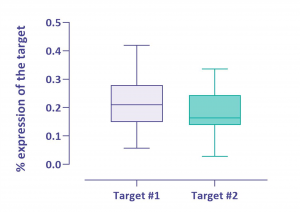
Two out of three targets were expressed at low levels in the cells. The graph shows the ability of the assays to quantify these low expression genes. More than 20 measurements were performed and all were analyzable despite this low representativity.
Clean Cells: a QC partner for development and testing
Clean Cells is a 20+ year quality control expert working with a wide array of biologics intended for humans and animals. Our portfolio of assays, now including dPCR options, has hundreds of assays for potency, safety, purity and identity evaluation.
Please reach out to our team to discuss testing needs or customized assay development.